When I first pulled MedGemma 4B into a clinical sandbox, my goal was simple: see if an open-weight model could survive the same red‑team prompts I usually reserve for commercial, closed LLMs. Within a weekend, I had it summarizing de‑identified oncology notes, critiquing draft radiology reports, and, just as important, clearly exposing where it still hallucinated.
In this MedGemma tutorial, I’ll walk through how I think about the model family as a health‑AI engineer: architectures, benchmarks, GPU sizing, deployment options, fine‑tuning strategy, and safety controls. The focus is practical: code‑ready guidance to help you de‑risk real clinical integrations under HIPAA/GDPR, not a glossy model launch recap.
Medical disclaimer: Nothing here is medical advice. MedGemma is a research/development tool (as of late 2025) and must not be used to make or replace clinical decisions without appropriate regulatory clearance, institutional review, and supervision by qualified clinicians.
Table of Contents
Understanding MedGemma: Google’s New Standard for Open Medical AI
MedGemma is Google Health’s open‑weight medical LLM line, released in 2025 as part of the Gemma ecosystem. It’s trained on a mix of biomedical literature, medical question‑answer pairs, and synthetic data, and it comes in both text‑only and multimodal variants.
From Med-PaLM to MedGemma: The Evolution of Medical LLMs
I think of Med-PaLM as the “proof of concept” and MedGemma as the “developer‑ready” successor. Med-PaLM and Med-PaLM 2 showed that large generalist models could hit clinician‑level performance on MedQA and similar exams, but they were closed and hard to integrate in regulated pipelines.
MedGemma keeps much of that medical reasoning focus but exposes:
- Open weights (see the official GitHub and Hugging Face repos) for inspection and on‑prem deployment.
- Smaller variants (4B, 27B) that are realistically runnable on hospital or cloud GPUs.
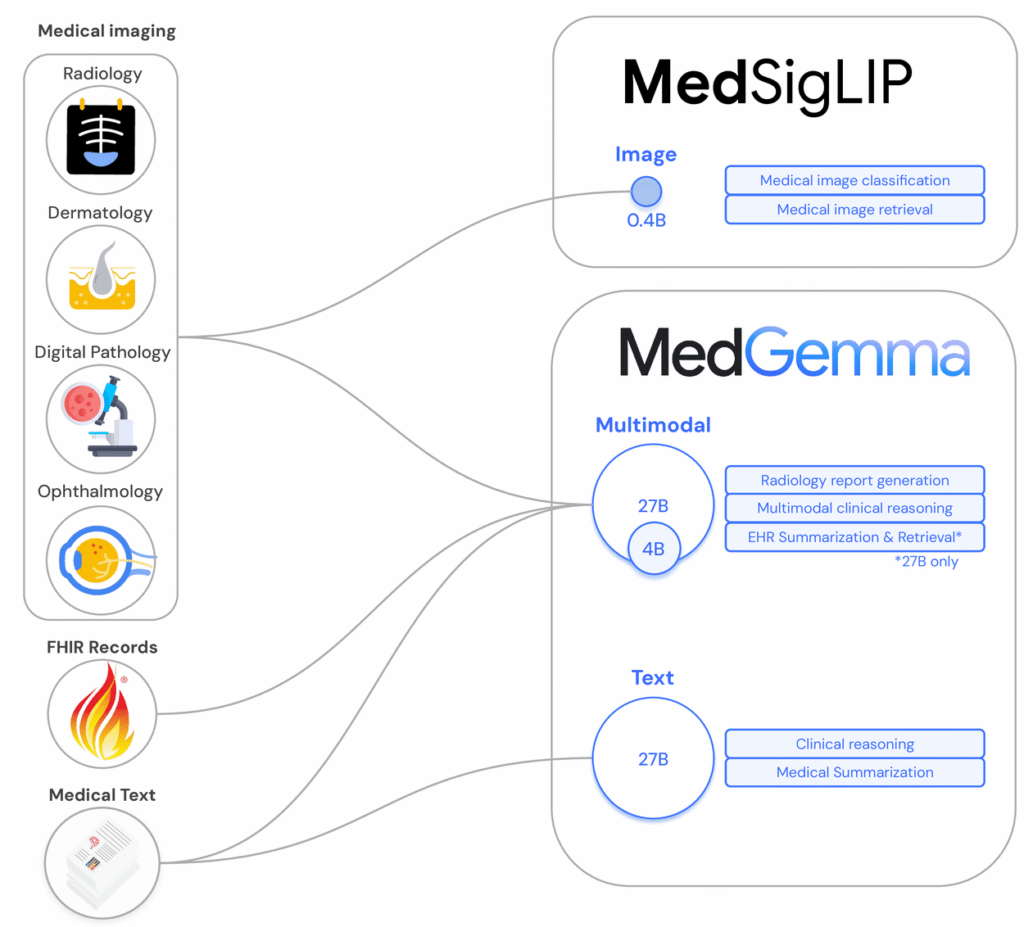
- Multimodal capabilities via MedSigLIP for imaging tasks like radiographs and pathology slides.
Open Weights vs. Closed API: Why MedGemma Matters
For regulated markets, open weights change the threat model:
- You can keep PHI in your own VPC or on‑prem cluster, avoiding cross‑border data transfer issues under HIPAA/GDPR.
- You can instrument and log everything, tokens, prompts, responses, for validation and incident review.
- You can fine‑tune and constrain the model using LoRA or adapters, rather than praying a vendor updates their safety filters.
The tradeoff is responsibility: you now own patching, access control, and safety evaluation. That’s exactly why a disciplined evaluation and deployment process is non‑negotiable with MedGemma.
Deep Dive: MedGemma Architectures & Benchmarks
MedGemma 4B vs 27B: Choosing Between Speed and Reasoning
MedGemma currently ships in two main instruction‑tuned text variants:
- medgemma-4b-it – ~4B parameters, efficient, good for latency‑sensitive pipelines, edge experimentation, or as a reranker/assistant around a larger model.
- medgemma-27b-text-it – ~27B parameters, noticeably stronger on long‑form reasoning, edge cases, and exam‑style questions.
In my testing on de‑identified endocrine consult notes, 4B worked for classification and summarization, while 27B added value for differential diagnosis explanation and guideline citation. If you’re building production decision support, I’d prototype with 4B but budget GPU for 27B.
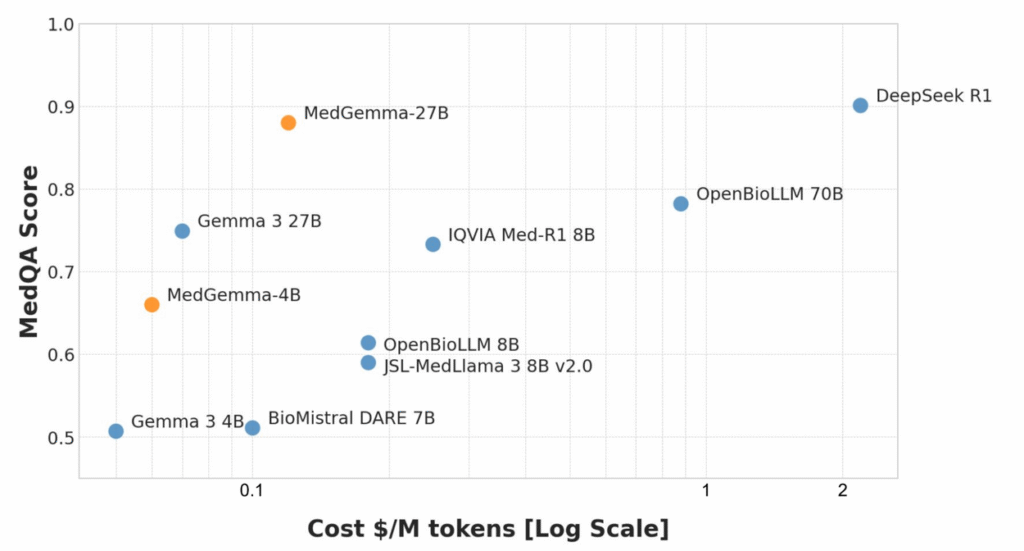
Performance Analysis: MedQA Scores and Clinical Accuracy
According to Google’s 2025 MedGemma paper and blog, 27B matches or exceeds earlier Med-PaLM‑class models on:
- MedQA (USMLE‑style) multiple‑choice exams
- MultiMedQA‑style composite benchmarks
- Several radiology and ophthalmology tasks (for multimodal variants)
Benchmarks give a ceiling: your real risk lies in distribution shift and hallucinations. I recommend:
- Building a local evaluation set of de‑identified cases mapped to guideline‑backed answers.
- Tracking factual accuracy, citation fidelity, and refusal rates per specialty.
- Logging adverse outputs (unsafe, biased, off‑label) and feeding them back into prompt/adapter tuning.
Inside the Tech: Multimodal Capabilities
Mastering Medical Imagery with MedSigLIP
MedGemma’s vision stack leverages MedSigLIP, a medical‑adapted variant of Google’s SigLIP. In practice, you pass an image embedding into the language model, then condition generation on both modalities.
In a radiology sandbox, I used the multimodal MedGemma to:
- Describe key findings on de‑identified chest X‑rays.
- Compare current vs prior images (e.g., nodule interval growth).
- Generate draft impressions for an attending to edit.
Two caveats:
- As of late 2025, these models are not FDA‑cleared devices: they’re research tools.
- I always required the model to output: “This is not a diagnostic report. A radiologist must review the images.”
Processing EHRs and Clinical Notes
Text‑only MedGemma works well on structured + free‑text EHR exports:
- Creating sectioned summaries (HPI, meds, labs, assessment/plan).
- Normalizing problems to SNOMED/ICD‑10 with a terminology layer on top.
- Flagging missing guideline‑recommended labs.
For PHI, I strongly prefer a pattern of: de‑identify → process with MedGemma → re‑link via pseudonyms inside your secure environment.
Developer Guide: How to Run MedGemma
Deployment Options: Hugging Face, Kaggle, and Vertex AI
You can get started three main ways:
- Hugging Face – Pull
google/medgemma-4b-itorgoogle/medgemma-27b-text-itviatransformersorvLLM. Good for on‑prem and VPC deployments. - Kaggle notebooks – Google’s example notebooks are handy for quick, free GPU prototyping (no PHI, obviously).
- Vertex AI – For GCP shops, Gemma/MedGemma integration lets you deploy managed endpoints, add IAM, and wrap with Cloud Logging and Policy Intelligence.
In my own hospital PoC, I used Hugging Face + vLLM on Kubernetes inside a private subnet and proxied requests through an audited API gateway.
Quick alternative for prototyping: Instant access to MedGemma via dr7.ai‘s unified API (free tier, no card needed).
Environment Setup: GPU Requirements for 4B and 27B
Very roughly (check official docs for updates):
- 4B: fits on a single 24 GB GPU (A10, L4) with 4‑bit quantization: great for dev and small batch inference.
- 27B: more comfortable on 2×24 GB or a single 80 GB GPU (A100/H100), especially for long sequences.
I recommend:
- Using vLLM or TensorRT‑LLM for production inference.
- Pinning container images with explicit CUDA/cuDNN versions.
- Adding resource limits and per‑tenant rate limiting to avoid denial‑of‑service and unexpected latency spikes.
Fine-Tuning for Specialized Healthcare Tasks
Step-by-Step: LoRA Fine-Tuning on Custom Medical Data
My default recipe for MedGemma specialization is parameter‑efficient fine‑tuning (PEFT) with LoRA:
- Curate de‑identified data: e.g., cardiology notes paired with guideline‑aligned summaries.
- Define a narrow task: “Summarize clinic notes for communication with PCPs,” not “general cardiology expert.”
- Use
peft+transformersto attach LoRA adapters to attention and MLP layers. - Train on instruction–response pairs with strong system prompts enforcing scope and disclaimers.
- Evaluate against a held‑out, clinician‑reviewed set before touching any real workflow.
I’ve seen LoRA adapters under 1–2% of base parameters meaningfully improve style, terminology, and alignment to local practice.
Data Privacy and De-identification Best Practices
For HIPAA/GDPR, I treat MedGemma as untrusted with raw PHI by default:
- Run a dedicated de‑identification pipeline (rule‑based + ML) before training or inference.
- Maintain data processing agreements and a RoPA (Record of Processing Activities).
- Never send rare disease case details to public endpoints or shared notebooks.
If you must work with limited PHI (e.g., on‑prem fine‑tuning), involve your IRB or data protection officer, document your safeguards, and apply strict access controls and audit logging.
Real-World Use Cases
Building an Automated Radiology Report Assistant
In one pilot, I connected multimodal MedGemma to a PACS sandbox with de‑identified chest X‑rays:
- The system ingested the image + brief clinical indication.
- MedGemma generated a structured draft (Findings, Impression, Recommendations).
- An attending radiologist edited and signed off.
The benefits were mostly consistency and speed for routine studies. Risks included occasional over‑confident descriptions of subtle findings. We mitigated this by forcing explicit uncertainty language and requiring radiologist sign‑off for every report. No direct patient‑facing outputs.
Clinical Decision Support Systems (CDSS)
For CDSS, I strongly prefer a retrieval‑augmented design:
- Retrieve relevant guidelines (e.g., ACC/AHA, NCCN) and local order sets.
- Let MedGemma summarize and contextualize those documents to the specific case.
- Log model rationales and expose them to clinicians for verification.
Any CDSS that influences orders or diagnoses will likely fall under medical device regulations (FDA, MDR), so you’ll need formal validation and post‑market surveillance, not just offline benchmarking.
Limitations and Responsible AI Usage
Handling Hallucinations in Medical Contexts
Even with strong MedQA performance, MedGemma will hallucinate:
- Fabricated citations or study names.
- Confident but wrong dosing or contraindications.
- Over‑generalization from adult to pediatric patients.
My mitigation toolkit:
- Constrain scope (education, summarization, documentation support, not autonomous diagnosis).
- Use RAG with authoritative sources (FDA labels, UpToDate‑style content, local protocols) and require the model to quote them.
- Add a second pass checker model or rules engine for high‑risk content (meds, labs, procedures).
- Make it impossible for the system to act without human confirmation in any safety‑critical path.
Google’s Safety Filters and Use Restrictions
Google’s documentation and licenses impose restrictions on:
- Certain high‑risk medical uses, especially autonomous diagnosis or treatment recommendations.
- Re‑identification attempts or misuse of PHI.
I treat Google’s guidance as a floor, not a ceiling. Internally, I overlay:
- A model governance process (model cards, risk assessments, approval gates).
- Regular safety audits with clinicians and security teams.
If a patient is acutely unwell, the correct instruction to any user of your system should be: “Call emergency services or contact your clinician immediately,” not “Ask the model.”
Conclusion: The Future of Open Source in Healthcare
MedGemma doesn’t magically solve clinical AI, but it finally gives us serious, open medical LLMs we can inspect, stress‑test, and deploy under our own compliance regimes.
My own pattern today is:
- Start with MedGemma 4B for low‑risk summarization and experimentation.
- Graduate to 27B for reasoning‑heavy tasks, always behind de‑identification and RAG.
- Layer LoRA adapters, governance, and strict human‑in‑the‑loop review for anything touching care.
Used thoughtfully, MedGemma can be one of those tools. Used recklessly, it’s just another way to hallucinate with higher confidence. The difference is in how you test, constrain, and govern it, and that part is entirely on us.
Disclaimer:
The content on this website is for informational and educational purposes only and is intended to help readers understand AI technologies used in healthcare settings. It does not provide medical advice, diagnosis, treatment, or clinical guidance. Any medical decisions must be made by qualified healthcare professionals. AI models, tools, or workflows described here are assistive technologies, not substitutes for professional medical judgment. Deployment of any AI system in real clinical environments requires institutional approval, regulatory and legal review, data privacy compliance (e.g., HIPAA/GDPR), and oversight by licensed medical personnel. DR7.ai and its authors assume no responsibility for actions taken based on this content.
MedGemma Tutorial – Frequently Asked Questions
What is MedGemma and who is this MedGemma tutorial for?
MedGemma is Google Health’s open‑weight medical large language model family, available in text‑only and multimodal variants. This MedGemma tutorial is aimed at health‑AI engineers, clinical informaticians, and technical teams who want practical, code‑level guidance for deploying MedGemma safely in HIPAA/GDPR‑constrained environments.
How do I choose between MedGemma 4B and MedGemma 27B for my project?
MedGemma 4B is smaller and faster, ideal for latency‑sensitive tasks like classification, summarization, or reranking. MedGemma 27B provides stronger long‑form reasoning, better for differential diagnoses explanations or guideline‑aware CDSS prototypes. A common pattern is prototyping with 4B, then allocating GPU budget for 27B in production.
How do I actually run MedGemma step‑by‑step?
You typically start by pulling the MedGemma model from Hugging Face, Kaggle, or deploying via Vertex AI. Set up GPUs (24 GB for 4B, larger or multiple GPUs for 27B), load with frameworks like transformers or vLLM, wrap it behind an API gateway, and add logging, access control, and rate limiting.
Can I use MedGemma for real clinical decision making today?
No. MedGemma is a research and development tool, not an FDA‑cleared medical device. It should not replace clinician judgment or be used for autonomous diagnosis, prescribing, or triage. Safe uses today focus on summarization, documentation assistance, education, and decision support with strict human review and regulatory oversight.
What are best practices for fine‑tuning MedGemma on my hospital’s data?
Use parameter‑efficient fine‑tuning (e.g., LoRA) on carefully curated, de‑identified instruction–response pairs for a narrow task. Run de‑identification before training, keep data on‑prem or in a secure VPC, involve your IRB or data protection officer, and evaluate against a clinician‑reviewed test set before any clinical pilot.
Past Review: