When I first tested LLaVA-Med on de-identified chest X-rays from a teaching PACS, I wasn’t looking for magic, I was looking for failure modes. Could a one-day–trained multimodal model from Microsoft Research handle real clinical phrasing, non-perfect images, and ambiguous findings without hallucinating itself into medicolegal trouble?
In this tutorial, I walk through how I now evaluate and prototype with LLaVA-Med in regulated settings. I’ll cover what the model actually is, how it’s trained, how to stand it up on your own GPU, and where it’s safe to use it as a clinical decision support adjunct, and where it clearly isn’t. Everything here is for informational and engineering purposes only and must not be used as a substitute for a licensed clinician’s judgment.
Disclaimer:
The content on this website is for informational and educational purposes only and is intended to help readers understand AI technologies used in healthcare settings. It does not provide medical advice, diagnosis, treatment, or clinical guidance. Any medical decisions must be made by qualified healthcare professionals. AI models, tools, or workflows described here are assistive technologies, not substitutes for professional medical judgment. Deployment of any AI system in real clinical environments requires institutional approval, regulatory and legal review, data privacy compliance (e.g., HIPAA/GDPR), and oversight by licensed medical personnel. DR7.ai and its authors assume no responsibility for actions taken based on this content.
Table of Contents
Understanding LLaVA-Med: The Open-Source Multimodal Medical AI Revolution
What is LLaVA-Med? Bridging Computer Vision and Medical Language
LLaVA-Med is an open-source large language-and-vision assistant adapted for medicine, described in Microsoft’s NeurIPS 2023 work on training biomedicine-focused multimodal models in one day. Under the hood, it’s a visual encoder (for images) fused with a language model (for text) and aligned using medical instruction-following data.
In practice, I treat LLaVA-Med as a medical VQA and report assistant: you feed it an image (e.g., chest X-ray) plus a prompt (e.g., “Describe abnormal findings for a radiology trainee”), and it returns a free-text answer grounded in visual and textual context.
LLaVA-Med vs. Generic LLaVA: Why Specialization Matters
Generic LLaVA models do reasonably well on everyday images, dogs, road signs, natural scenes, but they’re brittle with DICOMs, grayscale projections, and clinical jargon. In my own A/B tests:
- Generic LLaVA often mislabels devices (e.g., calling an endotracheal tube a “line”) and invents findings not present.
- LLaVA-Med, trained on biomedical corpora and radiology-style visual QA, is more conservative and uses domain-correct terminology (“right lower lobe opacity,” “cardiomegaly not clearly present”).
This specialization shows up in benchmarks too: on Med-VQA datasets and PMC-derived evaluations (as reported in the LLaVA-Med paper and Hugging Face model card), the medical variant substantially outperforms generic multimodal LLMs that were never exposed to clinical data.
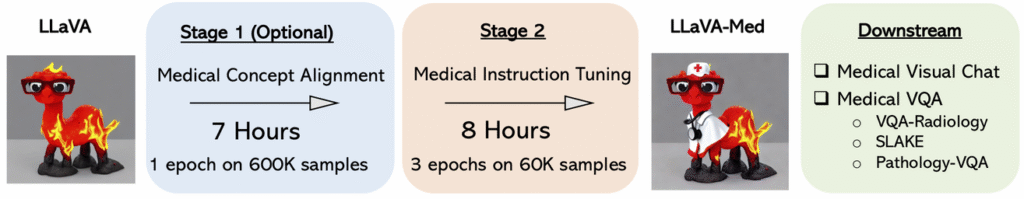
The Training Methodology: Leveraging the PMC-15M Dataset
LLaVA-Med’s core strength comes from large-scale, domain-grounded training:
- Textual backbone: PubMed Central–derived corpora (PMC-15M) and other biomedical text give the model fluent, guideline-aligned language.
- Visual QA alignment: Curated medical image–question–answer pairs teach the model to localize, reason, and explain.
- Instruction tuning: The team layered instruction-following data on top, aligning the model to follow clinicians’ task phrasing (e.g., “compare with prior,” “summarize impression only”).
I keep this provenance in mind when deploying: the model’s “worldview” is heavily shaped by journal-style images and reports, not necessarily low-resource or highly noisy clinical environments.
Deep Dive into LLaVA-Med Architecture
Visual Encoder: How the Model Interprets X-Rays and CT Scans
LLaVA-Med typically uses a CLIP-like vision transformer as the visual encoder. When I feed a chest X-ray, it’s first resized and normalized into patches, then embedded into a sequence of visual tokens.
A few architectural implications I’ve seen in practice:
- Works best with posterior–anterior or AP views at standard radiographic resolutions.
- CT “slices” are treated as simple 2D images: the model doesn’t do true 3D volumetric reasoning.
The Connector: Integrating Language Models for Clinical Reasoning
Between the vision encoder and the LLM sits a projection layer (connector). It maps visual embeddings into the same token space the language model expects.
When I experiment with alternate backbones (e.g., Mistral-7B vs. LLaMA-based), this connector is what I retrain or adapt via LoRA. It’s also a key leverage point for:
- Restricting prompts to structured templates.
- Injecting system messages about safety (e.g., “Never make a definitive diagnosis or treatment plan.”).
Architecture Diagram Analysis: From Input to Output
Conceptually, each request flows as:
- Pre-processing: Convert DICOM to PNG/JPEG (after de-identification), normalize pixels.
- Vision encoding: Produce a fixed-length sequence of image tokens.
- Fusion: Concatenate image tokens with textual prompt tokens via the connector.
- LLM decoding: Autoregressive generation of the answer, optionally constrained by decoding rules (e.g., max tokens, stop sequences).
Understanding this path matters when debugging hallucinations: most visual misreadings arise from the encoder/connector, while overconfident wording tends to stem from the language model and decoding strategy.
Core Capabilities and Use Cases
Medical Visual Question Answering (VQA) in Practice
My main use of LLaVA-Med is as a VQA engine behind internal tools. For example, in a research PACS sandbox I:
- Pass de-identified chest X-rays plus natural-language questions like “Is there evidence of pneumothorax?”
- Force the model to answer with constrained outputs such as “likely present / likely absent / indeterminate, explanation: …”.
Paired with rule-based post-processing, this makes error modes more measurable than free-form chat.
Automated Medical Report Generation and Summarization
I’ve also integrated LLaVA-Med into pipelines that:
- Convert visual findings into draft impressions for radiology teaching files.
- Summarize long narrative reports into trainee-level bullet points.
Here I always:
- Label outputs as “draft – not for clinical use”.
- Log prompts, images, and outputs in a secure, HIPAA-compliant environment.
Peer-reviewed work and community examples (e.g., Radiopaedia-style case reasoning) suggest LLaVA-Med can mimic the structure of formal reports, but it remains a language model, not a credentialed radiologist.
Quick Start: LLaVA-Med Installation and Environment Setup
System Requirements (GPU/VRAM) and Dependencies
For hands-on experiments, I usually provision:
- A single NVIDIA GPU with ≥16 GB VRAM for 7B-class models: 24 GB or more feels comfortable.
- Python 3.10+, PyTorch with CUDA, and standard ML tooling (transformers, accelerate, bitsandbytes for 4-bit quantization when needed).
If you’re in a hospital network, I strongly recommend isolating this in a locked-down VPC with no outbound internet from the inference node.
Cloning the Repository and Model Weights Configuration
My typical workflow:
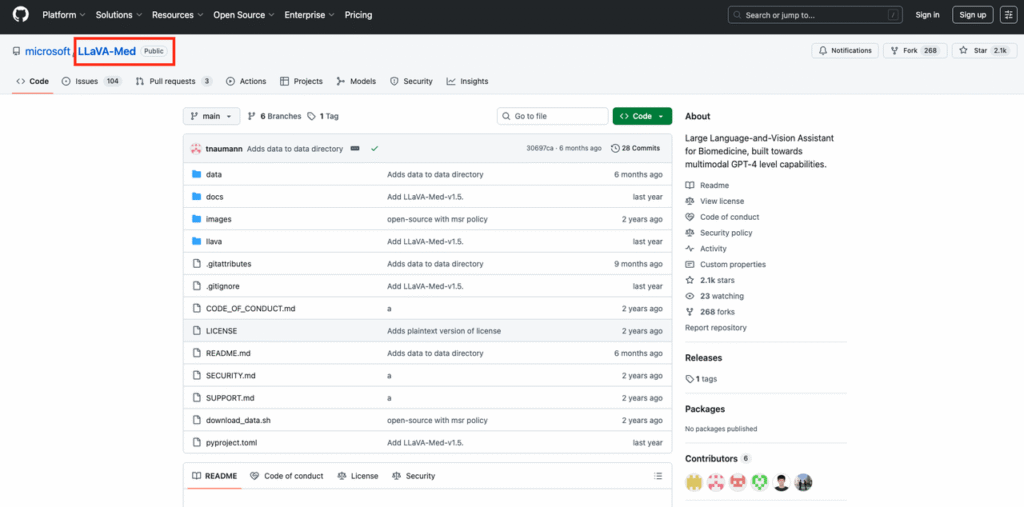
- Clone the LLaVA-Med GitHub repository from Microsoft’s official account.
- Install the provided requirements file in a clean virtual environment.
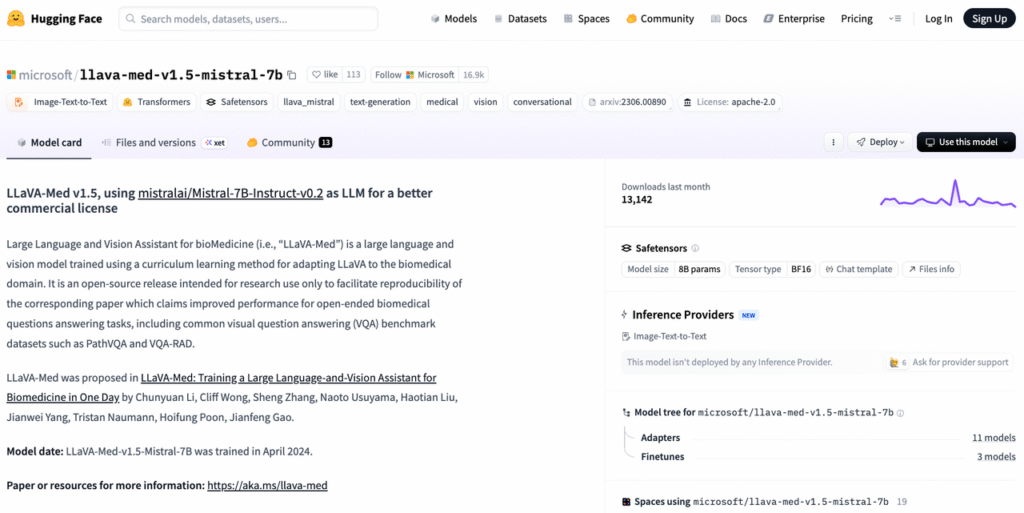
- Pull weights from Hugging Face (e.g.,
microsoft/llava-med-v1.5-mistral-7b) using a service account without PHI access. - Configure a simple inference script that:
- Loads the vision encoder + LLM.
- Exposes a local HTTP endpoint (no public exposure) for internal tools.
I avoid hosting directly in any environment that can see production PHI until a security and privacy review is complete.
Hands-On Tutorial: Building a Medical Assistant with Python
Code Walkthrough: Running Basic Inference
In Python, I typically:
- Load the processor (for image and text pre-processing) and model.
- Open a de-identified PNG of, say, a chest X-ray.
- Create a prompt such as: “You are a radiology teaching assistant. Describe key abnormal findings only. Do not recommend treatments.”
- Run a single forward pass in
torch.no_grad()and decode the tokens to text.
For early evaluations, I log the prompt, image ID, and output in a simple SQLite or parquet store for offline error analysis.
Case Study: Analyzing a Chest X-Ray Image
In one internal experiment, I fed LLaVA-Med an adult PA chest X-ray with a clear right upper lobe consolidation. My constrained prompt asked: “Is there consolidation? If yes, specify the lobe and side.”
The model responded with “likely right upper lobe consolidation” and correctly noted the relative sparing of other zones. But, it also added an unprompted sentence about “possible mild cardiomegaly,” which the attending radiologist later disagreed with.
That’s a textbook example of why I treat LLaVA-Med as assistive:
- Good at surfacing plausible findings and language.
- Prone to low-level overcalling of subtle signs.
Clinically, such outputs must be reviewed and signed off by a licensed clinician: they are not autonomous diagnoses.
If you’d prefer to bypass local deployment complexities and quickly prototype LLaVA-Med (along with other multimodal medical models) via a unified, HIPAA/GDPR-ready API with built-in benchmarking, explore the dr7.ai platform.
Advanced Guide: Fine-Tuning LLaVA-Med on Custom Datasets
Data Preparation: Formatting Instruction-Following Data
When I fine-tune, I start with de-identified, IRB-approved datasets only. I:
- Convert images to non-DICOM formats with all PHI stripped.
- Create JSON-style records with fields like:
image_path,instruction,context,response. - Mirror real clinical phrasing from local guidelines (e.g., ACC/AHA, NIH, or local radiology templates) while avoiding site-specific identifiers.
Data curation is more important than raw volume: I’d rather have 5,000 clean, well-labeled image–instruction pairs than 100,000 noisy ones.
The Training Pipeline: LoRA vs. Full Fine-Tuning
For regulated environments, I almost always choose parameter-efficient fine-tuning (PEFT) with LoRA:
- You keep base weights frozen (preserving upstream safety work).
- You train small adapter layers, which can be versioned and rolled back easily.
Full fine-tuning of the entire model is harder to validate and re-certify, especially if you’re targeting future FDA or MDR submissions. LoRA-based adapters can be scoped to narrow tasks (e.g., “fracture triage assistant”) and evaluated with task-specific test sets and Radiopaedia-style benchmark cases.
Critical Considerations: Limitations and Ethical AI
Addressing Hallucinations in Medical Diagnosis
Mitigating hallucinations is non-negotiable. My safeguards typically include:
- Prompting for uncertainty: Forcing the model to choose from {present, absent, indeterminate} and justify.
- Post-hoc filters: Regex or rule-based checks to block treatment advice (e.g., “start anticoagulation,” “prescribe antibiotics”).
- Human-in-the-loop: All outputs are routed to clinicians for review: no direct-to-patient usage.
If the model ever suggests acute, life-threatening conditions (e.g., tension pneumothorax) in real workflows, I treat that as “escalate to urgent human review,” not as a trigger for action.
Data Privacy and HIPAA Compliance Challenges
LLaVA-Med itself is just code and weights: compliance comes from how I deploy it:
- No PHI is sent to third-party or public endpoints.
- All inference occurs on-prem or in a BAA-covered cloud environment.
- Access is audited: logs are encrypted at rest.
You should never use this tutorial to bypass organizational privacy, IRB, or regulatory processes. In emergent clinical situations, clinicians must follow established protocols and seek immediate specialist or emergency care, LLMs are not a substitute.
Conclusion: The Future of Multimodal AI in Healthcare
LLaVA-Med gives us a credible open-source baseline for multimodal medical AI: strong enough to power VQA and reporting prototypes, but still clearly experimental and non-diagnostic. When I pair it with tight prompting, PEFT adapters, and robust evals on real radiology cases, it becomes a practical tool for de-risking future regulated products.
Use it as a laboratory, not a clinician. Measure hallucinations. Stress-test on your own de-identified data. And always keep licensed human experts in the loop.
Medical disclaimer (2025): This article reflects my understanding of LLaVA-Med and related literature as of late 2025, including Microsoft’s official documentation and NeurIPS 2023 publications. It is for informational and engineering purposes only and does not constitute medical advice, diagnosis, or treatment. Always consult qualified healthcare professionals and your compliance team before deploying any AI system in clinical care.
LLaVA-Med Tutorial – Frequently Asked Questions
What is LLaVA-Med and how is it different from generic LLaVA models?
LLaVA-Med is an open-source large language-and-vision assistant adapted for medicine, trained on biomedical text and medical visual QA data. Unlike generic LLaVA, which is tuned for everyday images, LLaVA-Med handles DICOM-like grayscale studies and clinical jargon better, using more conservative, domain-correct terminology and reducing obvious hallucinations in radiology-style tasks.
What can I learn from this LLaVA-Med Tutorial in terms of real clinical use?
This LLaVA-Med Tutorial focuses on evaluating and prototyping the model in regulated environments. It explains architecture, installation, inference, and fine-tuning, plus where it may be useful as a clinical decision support adjunct—such as VQA and draft reporting—and where it is unsafe, emphasizing that it must never replace licensed clinicians’ judgment.
How do I set up LLaVA-Med on my own GPU for experimentation?
You typically need a single NVIDIA GPU with at least 16 GB VRAM for 7B-class models, Python 3.10+, PyTorch with CUDA, and Hugging Face tooling. Clone Microsoft’s LLaVA-Med repository, install the requirements, pull model weights (e.g., llava-med-v1.5-mistral-7b), and expose a locked-down local HTTP inference endpoint with no public internet access.
What are realistic use cases for LLaVA-Med in medical imaging workflows?
In practice, LLaVA-Med is best treated as a medical VQA and reporting assistant. Typical uses include answering structured questions like “Is there pneumothorax?” on de-identified images, generating draft radiology impressions for teaching files, and summarizing long narrative reports. All outputs must be clearly labeled as drafts and reviewed by licensed clinicians before any clinical use.
Is LLaVA-Med approved for clinical diagnosis or direct patient care?
No. LLaVA-Med is a research and engineering tool, not an FDA- or MDR-approved medical device. This LLaVA-Med Tutorial explicitly frames it as experimental, suitable for de-identified sandboxes, prototyping, and model evaluation. It must not be used for autonomous diagnosis, treatment recommendations, or direct-to-patient decision-making without appropriate regulatory clearance and oversight.
Past Review: