In the intersection of artificial intelligence and healthcare, the ability to understand and process complex clinical text is paramount. Clinical notes, discharge summaries, and pathology reports are treasure troves of unstructured data that hold critical insights into patient health. ClinicalBERT emerged as a landmark model, specifically engineered to unlock this potential by adapting the powerful Transformer architecture to the unique language of medicine.
Table of Contents
The Genesis and Architecture of ClinicalBERT
ClinicalBERT is a specialized language model based on Google’s BERT (Bidirectional Encoder Representations from Transformers) architecture. Introduced by Kexin Huang, Jaan Altosaar, and Rajesh Ranganath in 2019, its primary innovation was its domain-specific pre-training. While the original BERT was trained on general text like Wikipedia and BookCorpus, ClinicalBERT was pre-trained on a massive, specialized corpus: the MIMIC-III (Medical Information Mart for Intensive Care III) dataset.
This dataset contains de-identified electronic health records (EHRs) from intensive care unit patients, providing a rich source of authentic clinical language. By training on an extensive collection of 1.2 billion words from diverse disease records, ClinicalBERT learned to grasp the nuances of medical terminology, abbreviations, and the often-unconventional sentence structures found in clinical notes. This domain-specific training allows it to build high-quality representations of medical concepts, a crucial advantage over general-purpose models.
The core idea was to create a model that could understand clinical text as proficiently as a medical professional, thereby enabling automated analysis for predictive tasks.
Core Applications and Downstream Tasks
The initial and most prominent application of ClinicalBERT was the prediction of 30-day hospital readmission. This is a significant challenge in healthcare, impacting patient quality of life and incurring substantial costs. The model was designed to dynamically assess a patient’s readmission risk by processing clinical notes written throughout their hospital stay, from early admission to the discharge summary.
As illustrated in the original paper, ClinicalBERT can update a patient’s risk score as new notes (e.g., nursing progress, radiology reports, physician notes) are added to their EHR. This provides clinicians with a dynamic tool to inform decisions and potentially intervene to prevent avoidable readmissions. In experiments, ClinicalBERT consistently outperformed baseline models like bag-of-words, bi-LSTMs, and even the original BERT on this task, highlighting the value of its specialized training.
Beyond readmission prediction, ClinicalBERT’;s flexible framework allows it to be fine-tuned for a wide array of “downstream” clinical tasks. Its powerful text representations have been successfully applied to:
- Mortality and Disease Prediction: Assessing the risk of mortality or predicting the onset of specific diseases based on note content.
- Medical Code Prediction: Automating the assignment of medical codes (like ICD codes) from unstructured notes, which is crucial for billing and research .
- Named Entity Recognition (NER): Identifying key entities in text such as diseases, symptoms, medications, and procedures.
- Relation Extraction: Uncovering relationships between entities, for example, detecting drug-drug interactions (DDIs) or linking a drug to an adverse event .
- Length-of-Stay Assessment: Predicting how long a patient is likely to remain in the hospital.
The Challenge of Long Clinical Notes and Evolving Solutions
A significant architectural limitation of the original BERT, and by extension ClinicalBERT, is its fixed maximum input length of 512 tokens (word pieces). This is a major hurdle in the clinical domain, where documents are often substantially longer. For instance, the average discharge summary in the MIMIC-III dataset is approximately 2000 words, which translates to nearly 3000 tokens, far exceeding the model’s capacity.
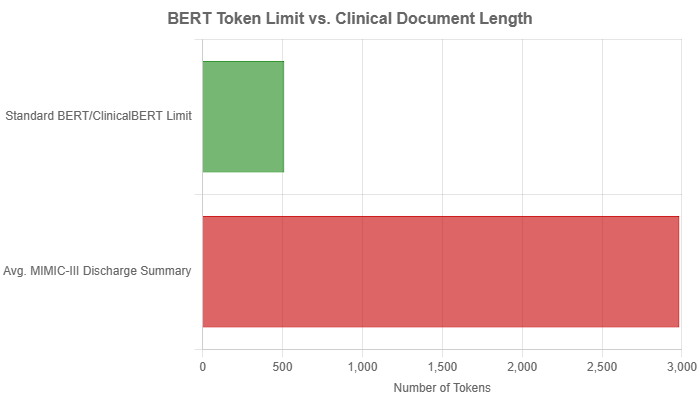
To work around this limitation, researchers developed several pre-processing strategies:
- Truncation: The simplest method, which involves cutting off the text at the 512-token mark. This can be done from the beginning (left truncation) or the end (right truncation), but it inevitably leads to information loss.
- Head-to-Tail: A more nuanced truncation strategy that combines the beginning and the end of a document. A common approach is to take the first 128 tokens and the last 382 tokens, preserving the introduction and conclusion, which are often information-rich .
- Hierarchical/Chunking: This method involves splitting the long document into smaller, overlapping chunks of 512 tokens. Each chunk is processed individually by the model, and the outputs are then aggregated (e.g., through mean or max pooling) to produce a final classification for the entire document.
While these workarounds enabled the use of ClinicalBERT on long documents, they are suboptimal as they fail to capture long-range dependencies that span across chunks. This limitation spurred the development of a new generation of models. Researchers introduced Clinical-Longformer and Clinical-BigBird, which use a sparse attention mechanism to extend the input sequence length to 4096 tokens. These models have demonstrated superior performance on tasks involving long clinical texts by being able to process entire documents at once, thereby capturing the full context.
Comparative Analysis: ClinicalBERT in the Landscape of Medical AI
ClinicalBERT did not exist in a vacuum. Its development and success can be best understood by comparing it to other models in the field.
vs. BlueBERT
Concurrent to ClinicalBERT, another model named BlueBERT was developed. While both are adaptations of BERT for the medical domain, they differ in their training data and primary evaluation focus.
- Training Data: ClinicalBERT was trained exclusively on clinical notes (MIMIC-III). In contrast, BlueBERT was pre-trained on a combination of PubMed abstracts and MIMIC-III notes. This gives BlueBERT a broader understanding of both biomedical research literature and clinical practice language.
- Evaluation: ClinicalBERT was primarily evaluated on the specific clinical task of readmission prediction. BlueBERT was evaluated on the broader Biomedical Language Understanding Evaluation (BLUE) benchmark, which includes diverse tasks like sentence similarity, NER, and document classification.
A direct performance comparison is difficult due to these differences, but both models unequivocally proved the benefit of domain-specific pre-training over general-purpose models.
vs. Long-Sequence Models
As mentioned, models like Clinical-Longformer and Clinical-BigBird were created to directly address the input length limitation of ClinicalBERT. Studies have shown that these long-sequence transformers consistently and significantly outperform ClinicalBERT on downstream tasks that rely on long documents. By processing up to 4096 tokens, they can learn long-term dependencies that ClinicalBERT misses, leading to new state-of-the-art results in clinical NLP.
Privacy and Ethical Considerations
Training language models on sensitive patient data from EHRs raises significant privacy concerns. A key question is whether Protected Health Information (PHI) could be “;leaked” or extracted from the trained model’s parameters. Researchers have actively investigated these risks.
Studies attempting to mount privacy attacks on BERT-based models have found that they are less susceptible to direct training data extraction compared to generative models like GPT-2. However, some research indicates a small risk from membership inference attacks, where an attacker could determine if a specific patient’s data was part of the training set. For example, a study by Lehman et al. (2021) explored these vulnerabilities by training BERT models on pseudo-re-identified MIMIC-III notes. While they were unable to meaningfully link a patient’s name to their medical conditions using simple methods, they acknowledged that more sophisticated techniques might succeed.
To mitigate these risks, automatic de-identification has become a critical step. This process involves using NLP to find and replace or remove personally identifiable information from clinical texts before they are used for model training. Research has shown that using an automatically de-identified corpus for domain adaptation does not negatively impact downstream performance, making it an effective strategy for balancing utility and privacy.
Conclusion
ClinicalBERT stands as a foundational contribution to clinical natural language processing. It decisively demonstrated that adapting large language models to the specific domain of medicine yields significant performance gains, paving the way for a new generation of powerful clinical AI tools. While its inherent limitation on input length has been addressed by newer architectures like Clinical-Longformer, the principles established by ClinicalBERT—the importance of domain-specific pre-training, the flexibility for diverse downstream tasks, and the need to confront privacy challenges—continue to shape the field. Its legacy is not just in its own performance, but in the wave of innovation and research it inspired, pushing the boundaries of what’s possible in automated healthcare analysis.

