In July 2025, Google introduced a significant advancement in medical artificial intelligence with the release of MedSigLIP, a collection of vision-language foundation models. The flagship open model, google/medsiglip-448, is a powerful encoder specifically designed to bridge the gap between medical imagery and textual descriptions. It represents a specialized variant of the SigLIP (Sigmoid Loss for Language Image Pre-training) architecture, fine-tuned to interpret the complex and nuanced data prevalent in healthcare.
This article provides a comprehensive analysis of google/medsiglip-448, exploring its architecture, training methodology, performance benchmarks, and practical applications, based on its official documentation and technical reports.
Table of Contents
Core Architecture and Technical Specifications
At its heart, MedSigLIP-448 is a dual-tower encoder model, a design proven effective for multimodal tasks. This architecture consists of two main components that work in parallel:
- Vision Encoder: A 400-million parameter Vision Transformer (ViT) responsible for processing and understanding visual information.
- Text Encoder: A 400-million parameter text transformer that processes and encodes textual data.
Together, these encoders map both images and text into a shared embedding space, allowing for direct comparison and calculation of similarity. This structure, totaling approximately 900 million parameters, is based on the SigLIP-400M architecture. Notably, this same vision encoder is the powerhouse behind the visual understanding capabilities of Google’s generative MedGemma models.
The model’s key technical specifications are tailored for a balance of performance and efficiency:
- Image Resolution: It processes images at a resolution of 448×448 pixels. This is a deliberate choice, offering a significant level of detail for medical images while remaining computationally more manageable than higher-resolution models.
- Context Length: The text encoder supports a context length of up to 64 tokens, sufficient for handling descriptive labels, captions, and short medical report phrases.
- Modalities: The model is inherently multimodal, accepting both images and text as input to produce embeddings or similarity scores.
This architecture makes MedSigLIP a lightweight yet powerful tool, suitable for deployment on edge devices and mobile platforms, as highlighted in a MarkTechPost article.
Training Methodology: A Hybrid Approach to Medical Specialization
The efficacy of MedSigLIP-448 stems from its unique training regimen, which can be described as a “;domain enhancement” process. Instead of being trained exclusively on medical data, the model was pre-trained on a vast and diverse dataset combining two types of data:
- De-identified Medical Data: A wide array of medical image-text pairs, including chest X-rays, dermatology images, ophthalmology images, histopathology slides, and slices from CT and MRI volumes, paired with their associated reports or descriptions.
- Natural Image Data: A collection of general, non-medical images and text pairs.
This hybrid approach is critical. The medical-specific data allows the model to learn the fine-grained features and terminology unique to clinical domains. Simultaneously, the inclusion of natural images ensures that the model retains its robust, general-purpose ability to parse and understand common visual concepts, preventing it from becoming overly specialized and brittle. This methodology, detailed in the model card, enables MedSigLIP to achieve high performance across various medical subfields while maintaining a broad understanding of the visual world.
Performance Benchmarks and Zero-Shot Capabilities
MedSigLIP’;s performance has been rigorously evaluated across several medical imaging tasks. One of the most compelling demonstrations of its power is in zero-shot classification of chest X-ray findings. In this setup, the model classifies images without any task-specific training, simply by comparing an image to a set of text prompts (e.g., “Cardiomegaly” vs. “Normal X-ray”).
The official documentation provides a direct comparison of MedSigLIP’s zero-shot Area Under the Curve (AUC) scores against ELIXR, another prominent model. The results are particularly impressive given that MedSigLIP operates on 448×448 images, while ELIXR uses a much higher 1280×1280 resolution.
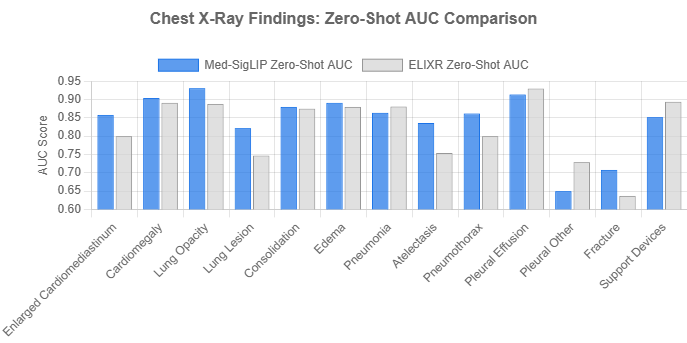
As the chart illustrates, MedSigLIP achieves an average AUC of 0.844, slightly outperforming ELIXR’s average of 0.824. It shows particularly strong performance in identifying ‘Lung Opacity’ (0.931 vs. 0.888) and ‘;Lung Lesion’ (0.822 vs. 0.747). This demonstrates the model’s high efficiency and its ability to capture critical diagnostic features even at a reduced image resolution, a key advantage for scalable applications.
The model’s performance suggests that the specialized medical pre-training effectively compensates for the lower input resolution, making it a highly data-efficient and computationally-aware solution for clinical tasks.
Practical Applications and Use Cases
Google recommends MedSigLIP for medical image interpretation applications that do not require text generation. For generative tasks, the companion MedGemma model is the preferred choice. The primary use cases for MedSigLIP-448 are:
1. Data-Efficient Classification
The pre-trained embeddings from MedSigLIP serve as a powerful starting point for training custom classifiers. Developers can generate embeddings for their medical images once and then use these fixed representations to train lightweight models for various classification tasks with very little labeled data and minimal computational overhead.
2. Zero-Shot Classification
As demonstrated in the performance section, the model excels at classifying images using only textual prompts. This is invaluable in scenarios where labeled training data is scarce or unavailable. By measuring the similarity between an image embedding and text embeddings for different classes (e.g., “a photo of an arm with a rash” vs. “a photo of an arm with no rash”), the model can perform classification on the fly.
3. Semantic Image Retrieval
The shared embedding space allows for powerful semantic search. A user can input a text query, such as “pleural effusion present,” and the model can rank a database of medical images based on how closely each image’s embedding matches the text query’s embedding. This is a transformative capability for medical research and clinical decision support.
4. Foundation for Fine-Tuning
MedSigLIP-448 provides a robust foundation for further specialization. Developers can fine-tune the entire model on a specific dataset to achieve state-of-the-art performance on a niche task. The existence of community-tuned models like medsiglip-448-ft-crc100k on Hugging Face attests to its adaptability.
Getting Started with MedSigLIP-448
Google has made MedSigLIP-448 highly accessible through platforms like Hugging Face and Google Cloud Model Garden. The Hugging Face Transformers library simplifies local use. Here is a conceptual overview of the workflow provided in the official documentation:
# 1. Load the model and processor from Hugging Face
from transformers import AutoProcessor, AutoModel
import torch
device = "cuda" if torch.cuda.is_available() else "cpu"
model = AutoModel.from_pretrained("google/medsiglip-448").to(device)
processor = AutoProcessor.from_pretrained("google/medsiglip-448")
# 2. Prepare images and text prompts
from PIL import Image
# (Image loading and resizing code)
imgs = [Image.open("image1.png").convert("RGB"), ...]
texts = ["a description of image 1", "another description", ...]
# 3. Process inputs and move to device
inputs = processor(text=texts, images=imgs, padding="max_length", return_tensors="pt").to(device)
# 4. Get model outputs
with torch.no_grad():
outputs = model(**inputs)
# 5. Interpret the results
# Similarity scores for zero-shot classification
logits_per_image = outputs.logits_per_image
probs = torch.softmax(logits_per_image, dim=1)
# Or get the raw embeddings for other tasks
image_embeddings = outputs.image_embeds
text_embeddings = outputs.text_embeds
For more detailed examples, Google provides Colab notebooks for both a quick start and for fine-tuning the model.
Conclusion
google/medsiglip-448 stands out as a meticulously engineered foundation model for medical vision-language understanding. By building on the robust SigLIP architecture and employing a sophisticated domain enhancement training strategy, it achieves an exceptional balance of specialized accuracy, generalist capability, and computational efficiency. Its strong zero-shot performance, particularly at a modest 448×448 resolution, underscores its potential to democratize access to advanced medical AI by lowering data and compute barriers.
As a key component of Google’s broader Health AI ecosystem, MedSigLIP provides developers and researchers with a versatile and powerful tool to accelerate the creation of next-generation healthcare applications, from data-efficient diagnostic aids to semantic search engines for clinical data.
For further reading, the primary academic source is the “MedGemma Technical Report,” available on arXiv.






