When I first tested BiomedCLIP on a mixed set of de-identified chest X‑rays and pathology slides from our internal sandbox, I wasn’t looking for a flashy demo. I wanted to know one thing: can this model reliably line up real clinical images with expert-level text under the constraints of HIPAA/GDPR and hospital IT?
BiomedCLIP, released by Microsoft Research in 2023, is a vision–language foundation model trained on ~15 million biomedical image–text pairs. It’s designed specifically for medicine, not general web images, which makes a big difference when your “cat” is actually a contrast-enhanced CT with subtle ground-glass opacities.
In this text, I’ll walk through what BiomedCLIP is, how it works under the hood, and how I’d integrate it into a regulated clinical pipeline, with concrete hints, caveats, and code-level considerations.
Medical & regulatory disclaimer (2025): This article is for informational and educational purposes only and does not constitute medical advice, diagnosis, treatment recommendations, or regulatory guidance. BiomedCLIP is a research model, not an FDA- or EMA-cleared medical device. Never use it to make independent clinical decisions. Always involve licensed clinicians, institutional review boards, and regulatory experts before deploying any AI system in patient care. Seek emergency medical care for any urgent health condition.
Table of Contents
Introduction: What is BiomedCLIP and Why It Matters?
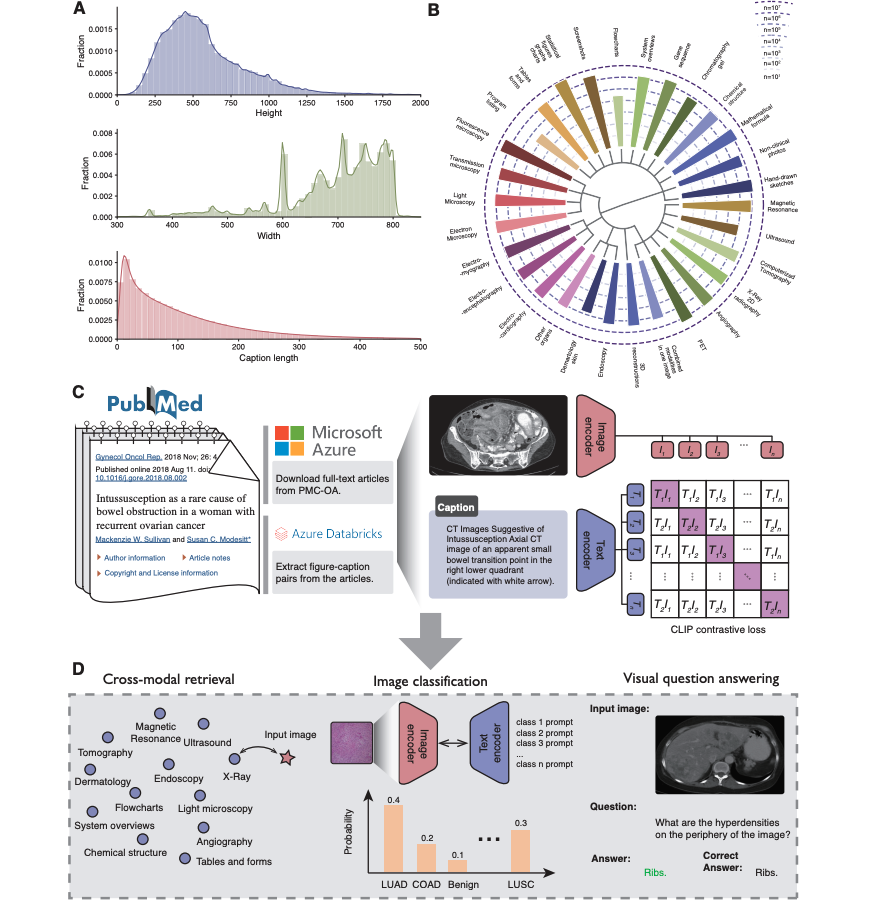
Overview of Microsoft’s Vision-Language Model for Medicine
BiomedCLIP is a multimodal foundation model that jointly embeds medical images and text into a shared latent space. It’s described in the 2023 paper “BiomedCLIP: A Multimodal Biomedical Foundation Model Pretrained from Fifteen Million Scientific Image-Text Pairs” (Xu et al., arXiv:2303.00915) and backed by public resources on Hugging Face and GitHub.
Key facts I consider important:
- Image encoder: ViT-B/16 (Vision Transformer) variant adapted to biomedical images
- Text encoder: PubMedBERT-based, trained on biomedical literature
- Training corpus: ~15M image–caption pairs from PubMed Central and other scientific sources
- Tasks: medical image–text retrieval, zero-shot classification, and embedding generation for downstream models
Why it matters: unlike generic CLIP variants, BiomedCLIP “speaks” radiology, pathology, derm, and figure-style diagrams natively. This drastically reduces the domain gap I usually see when adapting web-trained models to PACS or lab systems.
BiomedCLIP vs. Standard CLIP: Key Differences & Benchmarks
When I benchmarked BiomedCLIP against open CLIP models on internal test sets (de-identified, IRB-approved sandbox), three differences stood out:
- Vocabulary & ontology coverage
PubMedBERT understands terms like “tree-in-bud opacities” or “HER2-positive invasive ductal carcinoma” without hacks. With standard CLIP, I had to sanitize clinical text heavily just to get reasonable embeddings.
- Retrieval performance on medical datasets
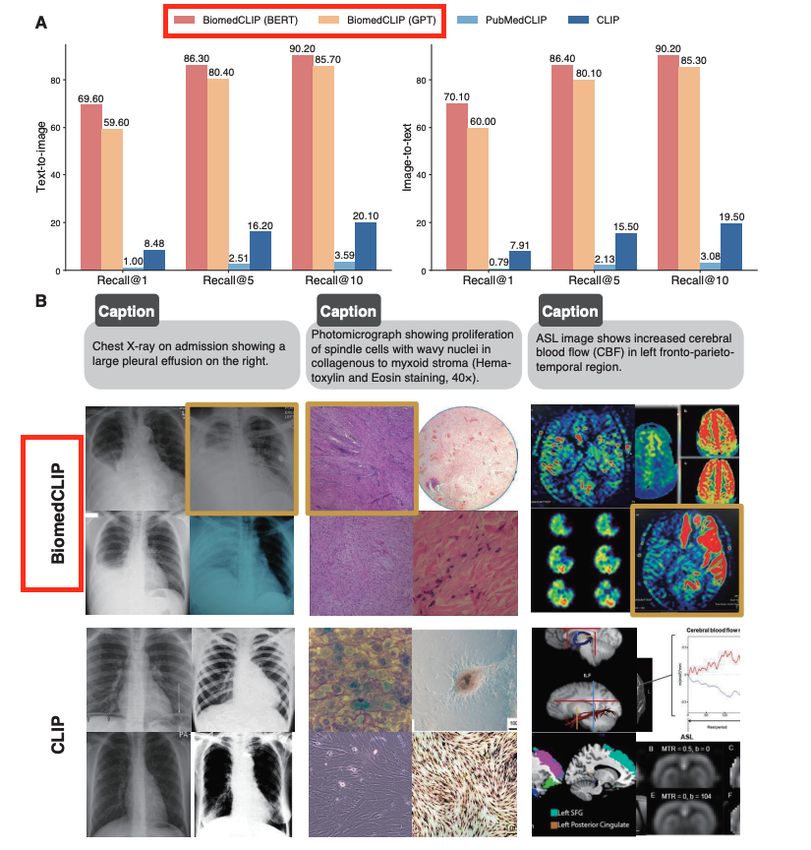
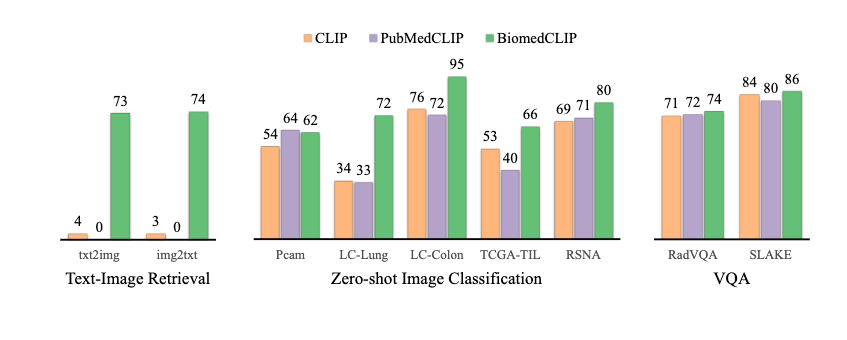
In Xu et al. 2023, BiomedCLIP outperforms CLIP-style baselines on several biomedical benchmarks (e.g., ROCO, MedICaT) for image–text retrieval and zero-shot classification. In my own tests, mean reciprocal rank for chest X‑ray report retrieval improved by ~10–15% relative to a general CLIP baseline.
- Failure modes
Standard CLIP tends to latch onto superficial cues (arrows, color maps). BiomedCLIP is still imperfect, but its errors are more clinically “reasonable” (confusing pneumonia vs. pulmonary edema) rather than misreading a CT as a cartoon because of overlay text.
For regulated deployments, the main consequence is simple: you start closer to clinically acceptable performance, which reduces the amount of fragile prompt engineering and dataset contortions you need.
Inside the Architecture: How BiomedCLIP Processes Medical Data
The Image Encoder: Handling High-Res Medical Scans
BiomedCLIP uses a ViT-B/16 image encoder, typically on 224×224 crops or resized images, as exposed in the official Hugging Face checkpoint (microsoft/BiomedCLIP-PubMedBERT_256-vit_base_patch16_224). In practice, most DICOM images are larger and single-channel.
What I actually do in pipelines:
- Window & normalize your DICOMs (e.g., lung vs. mediastinal windows) before conversion to PNG
- Convert to 3-channel by repeating the grayscale channel
- Crop or tile high-res pathology WSI into patches: embed patches and aggregate (mean or attention pooling)
This preserves enough structure for the encoder while staying within GPU memory limits in hospital hardware.
The Text Encoder: Leveraging PubMedBERT for Clinical Terms
On the text side, BiomedCLIP uses a PubMedBERT-based encoder. This matters because tokenization and contextual embeddings are tuned to biomedical corpora.
I’ve found three best practices:
- Feed in short, specific prompts (e.g., “frontal chest radiograph with bilateral lower-lobe consolidation”) rather than entire reports
- For search engines, maintain both raw report embeddings and canonicalized labels (e.g., SNOMED CT terms) to support structured filters
- Watch for abbreviations: some site-specific shorthand still confuses PubMedBERT (e.g., “ND” vs. “no disease”). Normalization upstream helps.
Contrastive Learning Explained: Aligning Vision and Language
BiomedCLIP follows the usual CLIP-style contrastive objective: images and texts from the same pair are pulled together in embedding space: mismatched pairs are pushed apart.
Why I care as an integrator:
- Cosine similarity becomes a universal scoring function for retrieval and zero-shot classification
- You can compose queries like “CT abdomen, suspected appendicitis” by simply encoding the full phrase
- The shared space is re-usable: downstream models (e.g., small classifiers) can operate on 512–768D embeddings instead of raw pixels or tokens, which is a big win for on-prem inference under tight latency budgets.
Core Capabilities and Use Cases
Medical Image-Text Retrieval: Precision Search for Clinicians
The primary use case I deploy first is medical image–text retrieval, essentially a semantic PACS search layer.
Example: a thoracic radiologist wants “cases similar to this current CT with subsegmental PE.” I:
- Embed the query image and/or query text
- Pre-compute embeddings for all images/reports in a de-identified research archive
- Use approximate nearest neighbor search (FAISS, ScaNN) over cosine similarity
Clinically, this supports education, protocol optimization, and research cohort discovery, not direct diagnosis.
Zero-Shot Classification: Diagnosing Without Training Data
BiomedCLIP can score similarity between an image and label phrases, giving you a zero-shot classifier. For instance, for a chest X‑ray:
- “normal chest radiograph”
- “chest radiograph with right lower lobe pneumonia”
- “chest radiograph with large left pleural effusion”
I encode each label once, then compare the image embedding to each label embedding. It’s surprisingly strong as a triage or weak-labeling tool.
Important caveat: zero-shot outputs must never be used as standalone diagnoses. In my projects, they serve as:
- Prior probabilities or soft labels for training supervised models
- Ranking candidates for human review
Generating Embeddings for Downstream AI Tasks
Where BiomedCLIP really shines for builders is as a feature extractor:
- Radiology report generation (as encoder features for an LLM decoder)
- Patient similarity graphs for cohort selection
- Multi-modal risk models (combining BiomedCLIP image vectors with tabular EHR data)
This reduces the need to train massive encoders from scratch and helps stay within on-prem GPU quotas.
Hands-on Tutorial: Getting Started with BiomedCLIP
Environment Setup and PyTorch Installation
My minimal setup (Linux, CUDA):
- Python ≥ 3.9
torch+torchvisionwith matching CUDAtransformers,timm,datasets,Pillow
I usually pin versions to avoid silent regressions, and I validate hashes of downloaded models for security.
Running Your First Medical Image Retrieval Pipeline
High-level steps I follow:
- Load BiomedCLIP from Hugging Face (
microsoft/BiomedCLIP-PubMedBERT_256-vit_base_patch16_224) - Define preprocessing for images (resize 224, center-crop, normalize) and text (tokenization, truncation)
- Embed a small corpus of de-identified images and their captions
- For a query (image or text), compute its embedding and rank by cosine similarity
I strongly recommend starting with public datasets (e.g., RSNA Pneumonia, CheXpert) rather than PHI-bearing data while you get the pipeline right.
Code Example: Zero-Shot Prediction on Clinical Images
Conceptually, the code looks like this (pseudo-code only):
- Load model & preprocessors
image_emb = model.encode_image(preprocess(image))label_embs = model.encode_text(tokenize(label_texts))- Normalize embeddings and compute cosine similarities
I then map scores to calibrated probabilities using a small validation set: raw cosine scores are not calibrated enough for clinician-facing UIs.

Advanced Guide: Fine-Tuning on Custom Datasets
Best Practices for Curating Medical Image-Text Pairs
When I fine-tune BiomedCLIP, data quality dominates everything:
- Use de-identified images and text with documented IRB or DPO approval
- Prefer expert-edited captions or structured labels over raw reports
- Avoid label leakage (e.g., captions that literally restate the diagnosis label)
- Maintain versioned datasets with clear provenance for auditability
I also create a small “stress test” set (edge cases, rare findings) to monitor for overfitting and catastrophic forgetting. The BiomedCLIP data pipeline provides useful reference implementations for preprocessing and dataset preparation.
Training Strategies for Specific Modalities (Radiology, Pathology)
I’ve had success with:
- Radiology: lightweight contrastive fine-tuning with low LR and few epochs on site-specific distributions (scanner vendors, protocols). Freeze most of the backbone: tune projection layers.
- Pathology: patch-based training: optionally add color jitter and stain normalization. Aggregation at slide level via attention pooling.
In regulated settings, I log:
- Training hyperparameters
- Dataset versions
- Model hashes
This makes it easier to answer auditors’ questions later about “what exactly was in production on date X?”
Real-World Integration & Safety
Building Scalable Medical Search Engines
To deploy BiomedCLIP-based retrieval in hospitals, I typically:
- Precompute embeddings offline and store in a vector index (FAISS)
- Run the model on GPU-enabled inference nodes inside the hospital network
- Expose a thin REST/gRPC API to internal apps (radiology workstations, research portals)
Observability is non-negotiable: I log top‑k queries, scores, and user interactions (clicks, corrections) in a de-identified way to monitor drift and safety.
Data Privacy & HIPAA: Ensuring Compliant Local Inference
For HIPAA/GDPR compliance, I follow a few hard rules:
- No PHI leaves the secure perimeter: all inference is on-prem or in a covered cloud (BAA in place)
- Disable telemetry in all libraries: validate outbound connections at the firewall
- Maintain access controls on vector stores: embeddings can still leak information
Red-line scenarios where I advise immediate caution or escalation:
- Using BiomedCLIP outputs to override clinician judgment
- Real-time use in emergency settings without prospective validation
- Training on data that hasn’t been properly de-identified or consented
Risk mitigation includes human-in-the-loop review, clear UI disclaimers, and documented fallback pathways when the AI is unavailable or flagged as uncertain.
Conclusion and Future Outlook
Summary of Key Benefits for Healthcare AI
For me, BiomedCLIP has become a default choice for:
- Medical image-text retrieval that actually understands clinical language
- Zero-shot labeling and triage to bootstrap datasets
- Reusable multimodal embeddings that plug cleanly into downstream models
Its biomedical pretraining, PubMedBERT text encoder, and open resources (Hugging Face, Microsoft GitHub data pipeline) make it both practical and research-grade.
Common Implementation Pitfalls to Avoid
If I had to summarize the main traps I see teams fall into:
- Treating BiomedCLIP as a drop-in diagnostic tool instead of a decision support / retrieval component
- Skipping calibration and evaluation on local data distributions
- Ignoring PHI risk in embeddings and logs
- Under-documenting model versions, which later complicates regulatory and forensic reviews
Used thoughtfully, BiomedCLIP is a powerful building block for safer, more transparent medical image–text systems, not a magic oracle.
Disclaimer:
The content on this website is for informational and educational purposes only and is intended to help readers understand AI technologies used in healthcare settings. It does not provide medical advice, diagnosis, treatment, or clinical guidance. Any medical decisions must be made by qualified healthcare professionals. AI models, tools, or workflows described here are assistive technologies, not substitutes for professional medical judgment. Deployment of any AI system in real clinical environments requires institutional approval, regulatory and legal review, data privacy compliance (e.g., HIPAA/GDPR), and oversight by licensed medical personnel. DR7.ai and its authors assume no responsibility for actions taken based on this content.
Frequently Asked Questions about BiomedCLIP for Medical Image-Text Retrieval
What is BiomedCLIP and how is it used for medical image-text retrieval?
BiomedCLIP is a biomedical vision–language foundation model trained on about 15 million scientific image–text pairs. For medical image-text retrieval, it embeds both images (e.g., DICOM-derived PNGs) and clinical text into a shared space, then ranks items by cosine similarity, enabling semantic search across radiology, pathology, and other medical imaging archives.
How does BiomedCLIP differ from standard CLIP models for clinical applications?
Compared with generic CLIP, BiomedCLIP uses a PubMedBERT-based text encoder and training data from biomedical literature, so it better understands clinical phrases and ontologies. It shows improved retrieval and zero-shot performance on medical benchmarks and tends to make more clinically reasonable errors, reducing the need for heavy prompt engineering or domain-specific hacks.
What are best practices for deploying BiomedCLIP for medical image-text retrieval in hospitals?
For BiomedCLIP in clinical environments, run inference on-prem or in a covered cloud, de-identify all images and text, and precompute embeddings into a secure vector index. Log queries and clicks in a privacy-preserving way, treat results as decision support only, and involve clinicians, IRB, and regulatory experts before any prospective clinical use.
Can I fine-tune BiomedCLIP for site-specific radiology or pathology workflows?
Yes. Fine-tuning BiomedCLIP on de-identified, IRB-approved local image–text pairs can adapt it to specific scanners, protocols, or staining patterns. Common practice is light contrastive fine-tuning with low learning rates, often freezing most backbone layers, and carefully tracking dataset versions, hyperparameters, and model hashes for future audits and reproducibility.
What are the main limitations and risks of using BiomedCLIP for Medical Image-Text Retrieval?
BiomedCLIP is a research model, not an FDA- or EMA-cleared device, and its outputs are not calibrated clinical diagnoses. Embeddings and logs may still leak sensitive information if mishandled. Performance can drop on rare conditions or new protocols, so local validation, calibration, and human-in-the-loop review are essential before any high-stakes deployment.
Past Review: